Dr. PCR BCR-ABL1 Major IS Detection Kit
CatalogOncologySolid Tumor

HNPCC (Hereditary non-polyposis colorectal cancer) is an autosomal dominant genetic condition predisposing to the early development of various cancers including those of colon, rectum, endometrium, etc. This disease is due to germline mutations in genes encoding proteins responsible for the repair of DNA replication errors, which are referred to as DNA mismatch repair (MMR) genes.
The microsatellite instability phenotype, characterized by widespread somatic alterations in the length of associated tumor and in 10-15% sporadic gastric and colon cancer.
HNPCC kit 1 – FL allows microsatellites instability analysis in tumor-associated HNPCC and in sporadic colon and stomach cancer by pentaplex PCR and then amplicon capillary electrophoresis by automatic sequencer.
L
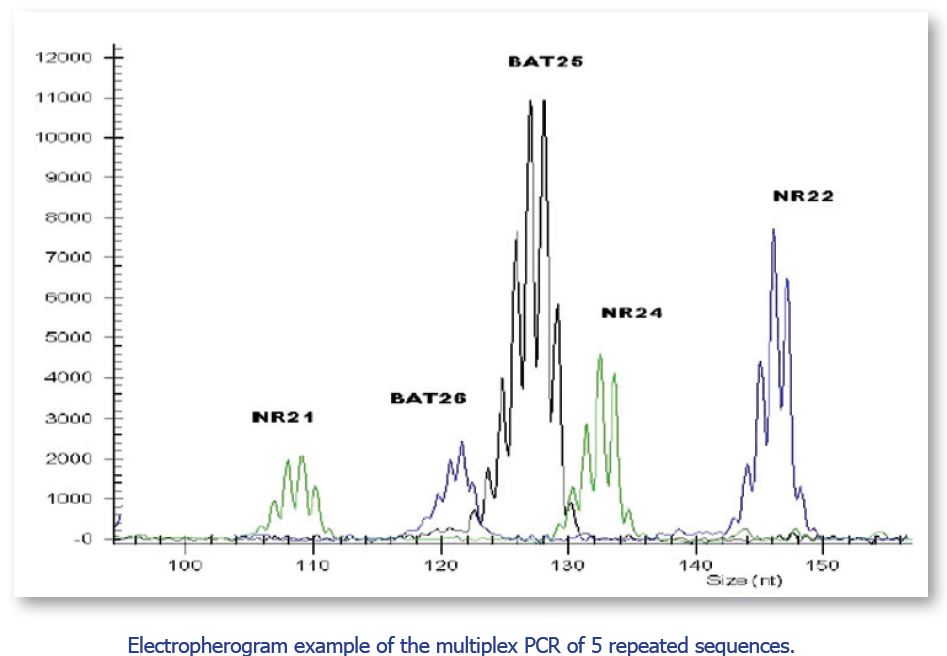
In the kit are provided five mononucleotide “almost-monomorphic” markers (BAT 25, BAT 26, NR21, NR22, NR24) dyed with fluorophores 6-FAM, HEX and TAMRA, for ABI Prism Instrument (Applied Biosystems) use.
| PRODUCT | QUANTITY |
|---|---|
| MS.01FL | 40 test /kit |
Public Documentation
Related products
2021 © Diagnóstica Longwood SL
This website uses cookies so that we can provide you with the best user experience possible. Cookie information is stored in your browser and performs functions such as recognising you when you return to our website and helping our team to understand which sections of the website you find most interesting and useful.
Strictly Necessary Cookie should be enabled at all times so that we can save your preferences for cookie settings.
If you disable this cookie, we will not be able to save your preferences. This means that every time you visit this website you will need to enable or disable cookies again.
This website uses Google Analytics to collect anonymous information such as the number of visitors to the site, and the most popular pages.
Keeping this cookie enabled helps us to improve our website.
Please enable Strictly Necessary Cookies first so that we can save your preferences!
Más información sobre nuestra política de cookies
